New Enzymes for Biocatalysis Created by the Wood Group
Epoxide Hydrolases
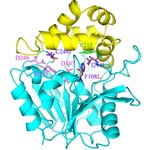
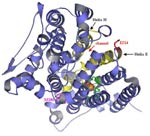
EChA. Epoxide hydrolase from Agrobacterium radiobacter AD1 triple mutant F108L/I219L/C248I, which has enhanced activity toward aerobic mineralization of cis-1,2-dichloroethylene. There are two domains of the epoxide hydrolase (EchA, Genbank accession number Y12804): the cap domain (yellow) and the core domain (light blue). The catalytic triad, Asp-107, Asp-246, and His-275, are pink. The proton-donor, Tyr-215, is green, the wild-type residues, Phe-108, Ile-219, and Cys-248 are blue, and the three amino acid substitutions of the triple mutant, F108L, I219L and C248I are red.
Epoxide hydrolase from Agrobacterium radiobacter AD1 triple mutant F108L/I219L/C248I.
Epoxide hydrolase from Agrobacterium radiobacter AD1 mutant I219F, which has enhanced enantioselectivity toward racemic styrene oxide for the production of (R)-1-phenylethane-1,2-diol. The catalytic triad residues Asp-107, Asp-246, and His-275 (pink) of the epoxide hydrolase are located between the core domain (light blue) and cap domain (yellow). Wild-type residue Ile-219 is shown in blue, and the amino acid substitution Ile-219-Phe is shown in red. Tyr-215 (green) is the proton-donor, which is in van der Waals contact with Ile-219.
Agrobacterium radiobacter AD1 mutant I219F animation.
Dioxygenases
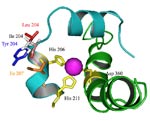
DNT DDO. 2,4-Dinitrotoluene (DNT) dioxygenase from Burkholderia sp. strain DNT with mutations I204L and I204Y in the DntAc (alpha subunit) enhances degradation toward 2,3-DNT, 2,4-DNT, 2,5-DNT, 2,6-DNT, 2-nitrotoluene (NT), and 4NT. Only a portion of an alpha subunit three-dimensional structure model is shown from the catalytic oxygenase component that consists of three alpha and three beta subunits (a3b3). Ile 204 of the wild-type DDO (white), variant DntAc I204L (red), and variant DntAc I204Y (blue) are shown. Residues His 206, His 211, and Asp 360 (yellow) coordinately bind with the mononuclear iron (pink sphere). Portions of the two ribbons of DDO which form the substrate channel are shown in light blue (terminating at Lys 190 and His 211) and green (terminating at Ala 343 and Ser 376). The orange residue (Ile 207) participates in the new hydrogen bonds (pink) with the mutated residues.
2,4-Dinitrotoluene (DNT) dioxygenase from Burkholderia sp. strain DNT with mutation I204L in the DntAc (alpha subunit) enhances degradation toward 2,3-DNT, 2,4-DNT, 2,5-DNT, 2,6-DNT, 2-nitrotoluene (NT), and 4NT. A three-dimensional model of a single alpha and beta subunit is shown from the catalytic oxygenase component that consists of three alpha and three beta subunits (a3b3). The DntAc mutation I204L is shown in red. Residues His 206, His 211, and Asp 360 (yellow) coordinately bind with the mononuclear iron (blue sphere). Helixes, sheets, and loops are shown in light blue, magenta, and pink, respectively.
Whole 1204L animation.
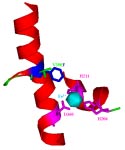
R34 DDO. 2,4-Dinitrotoluene dioxygenase from Burkholderia cepacia R34 with mutation V350F (V350 is green, F350 is blue) in the alpha subunit of the oxygenase large subunit, DntAc. This mutation enhances the oxidation of the green chemistry compounds o-methoxyphenol, o-cresol and o-nitrophenol. The DntAc residues H206, H211, and D360 (pink) are the coordinating residues of the mononuclear iron (light blue) of the DntAc active site. These metal binding residues, in addition to the residue at position 350, form the active site pocket of the terminal oxygenase.
2,4-Dinitrotoluene dioxygenase from Burkholderia cepacia R34 with
mutation V350F in the alpha subunit of the oxygenase large subunit, DntAc animation.
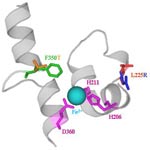
U2 DDO. Naphthalene dioxygenase from Ralstonia sp. strain U2 showing saturation mutagenesis variant NagAc F350T (F350 is green and T350 is orange), which has increased oxidation of 2,6-dinitrotoluene and 2-amino-4,6-dinitrotoluene, and DNA shuffling variant NagAc L225R (L225 is red and R225 is blue), which has increased activity towards 2,3-dinitrotoluene and 4-amino-2-nitrotoluene. Mononuclear iron (light blue) is coordinated by H206, H211, and D360 (magenta).
U2 DDO animation.
Monooxygenases
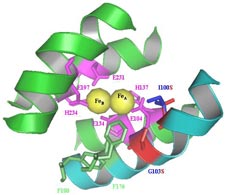
ToMO. Catalytic residues M180 and E214 of the hydroxylase alpha subunit (TouA) of toluene-o-xylene-monooxygenase from Pseudomonas stutzeri OX1 are important for enzyme activity for methyl and nitroaromatics oxidation as well as chlorinated ethene degradation. The C-terminal loop of helix E (yellow) and the N-terminal loop of helix H (yellow) form a channel at the surface of TouA. TouA M180 is pink and TouA E214 is red. Side chains shown in green are the metal-binding residues forming the diiron center (TouA-E104, E134, H137, E197, E231, H234)
The alpha subunit (TouA) of the toluene-o-xylene monooxygenase (ToMO) hydroxylase from Pseudomonas stutzeri OX1 is visualized here to show the catalytic residues M180 and E214. The ToMO hydroxylase has two each of the alpha, gamma, and beta subunits. The C-terminal loop of helix E (yellow) and the N-terminal loop of helix H (yellow) form a channel at the surface of TouA. TouA M180 is pink and TouA E214 is red. Side chains shown in green are the diiron-binding residues forming the catalytic center (TouA-E104, E134, H137, E197, E231, H234). Position E214 and M180 are important for enzyme activity for methyl and nitroaromatics oxidation as well as chlorinated ethene degradation.
Tomo hydroxylase animation.
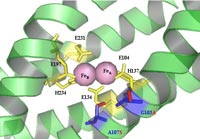
The alpha subunit (TouA) of the toluene-o-xylene monooxygenase (ToMO) hydroxylase from Pseudomonas stutzeri OX1 is visualized here using Pymol program to show the locations of the important hydroxylase residues of toluene monooxygenases. The ToMO hydroxylase has two each of the alpha, gamma, and beta subunits. The diiron center is shown in orange. Many of the beneficial positions such as TouA I100 (blue), A101 (yellow), E 103 (orange), A107 (red), and A110 (green) are located in the TouA B-helix (highlighted in red). Other beneficial positions such as T201 (purple), F205 (light blue), and E214 (pink) are located in the TouA E-helix (highlighted in yellow). Position Q141 and M180 are shown in light pink and dark pink, respectively. Many of these positions are important for influencing enzyme activity and regiospecificity.
The alpha subunit (TouA) of the toluene-o-xylene
monooxygenase (ToMO) hydroxylase from Pseudomonas stutzeri OX1 animation.
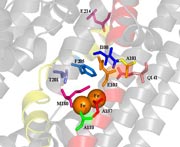
TpMO. Toluene para-monooxygenase from Ralstonia pickettii PKO1 TbuA1 double mutant I100S/G103S, the first truly meta-hydroxylating toluene monooxygenase. The TpMO hydroxylase consists of two alpha, two beta, and two gamma subunits, and only part of the alpha (TbuA1) subunit is shown. Wild-type residues shown in blue and mutated residues in red. Six coordinating residues anchoring the diiron-binding sites (yellow spheres) are shown in magenta. The four-helix bundle of TbuA1 anchoring the diiron-binding sites is shown in green and light blue.
Toluene para-monooxygenase from Ralstonia
pickettii PKO1 TbuA1 double mutant I100S/G103S animation.
T4MO. Toluene 4-monooxygenase from Pseudomonas mendocina KR1 TmoA double mutant G103A/A107S, altering o-methoxyphenol oxidation product from 4-methoxyresorcinol to 3-methoxycatechol. The T4MO hydroxylase consists of two alpha, two beta, and two gamma subunits, and only part of the alpha (TmoA) subunit is shown. Wild-type residues shown in blue and mutated residues in red. Six coordinating residues anchoring the diiron-binding sites (pink spheres) are shown in yellow. The four-helix bundle of TmoA anchoring the diiron-binding sites is shown in green.
Toluene 4-monooxygenase from Pseudomonas mendocina
KR1 TmoA double mutant G103A/A107S animation.
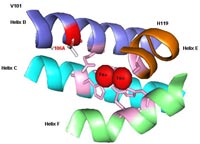
TOM. Toluene ortho-monooxygenase from Burkholderia cepacia G4 with mutation V106A (in red) in the alpha subunit (TomA3). This mutation enhances 1-naphthol synthesis and chlorinated ethene degradation. This is the first directed evolution of a non-heme monooxygenase. Portions of the four-helix bundle (helix B, helix C, helix E, and helix F) of TomA3 are shown. The six coordinating residues (E110, E140, H143, E201, E235, and H238) are shown in pink and anchor the diiron centers (red spheres). The smaller side chain (V106A) allows larger substrates to enter the active site; hence residue V106 is a gate.
Toluene ortho-monooxygenase from Burkholderia cepacia
G4 with mutation V106A (in red) in the alpha subunit (TomA3) animation.
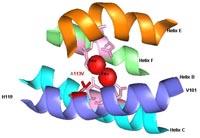
Toluene ortho-monooxygenase from Burkholderia cepacia G4 with one mutation A113V (in red) in the alpha subunit (TomA3). The mutant produces indigo from indole and turns blue in LB agar plates, while wild-type produces brown isoindigo. Portions of the four-helix bundle (helix B, helix C, helix E, and helix F) of TomA3 are shown. The six coordinating residues (E110, E140, H143, E201, E235, and H238) are shown in pink and anchor the diiron centers (red spheres). Saturation mutagenesis at TomA3 position 113 created various colored mutants due to the production of isatin (A113H), indirubin (A113F, A113I, and A113S), and unknown yellow compounds (A113G) from indole.
Toluene ortho-monooxygenase from Burkholderia cepacia
G4 with one mutation A113V (in red) in the alpha subunit (TomA3) animation.